The end of GSoC 2014 coding phase is getting closer and closer. This week I’m completing the tasks listed in the previous post.
Tasks completed this week:
Normalizing constants in 2d/3d basis functions
It came out that 2D gaussian basis function in the previous KCSD script was not normalized. After normalization the tests which compare KCSD2D to matlab scripts are not passing. Unwillingly, I’m dropping the old KCSD2D comparison tests.
Creating validation framework
This task is about creating a set of routines which help to compare 1D, 2D, 3D KCSD reconstruction against model CSD and potentials calculated using forward scheme.
Sample reconstructions
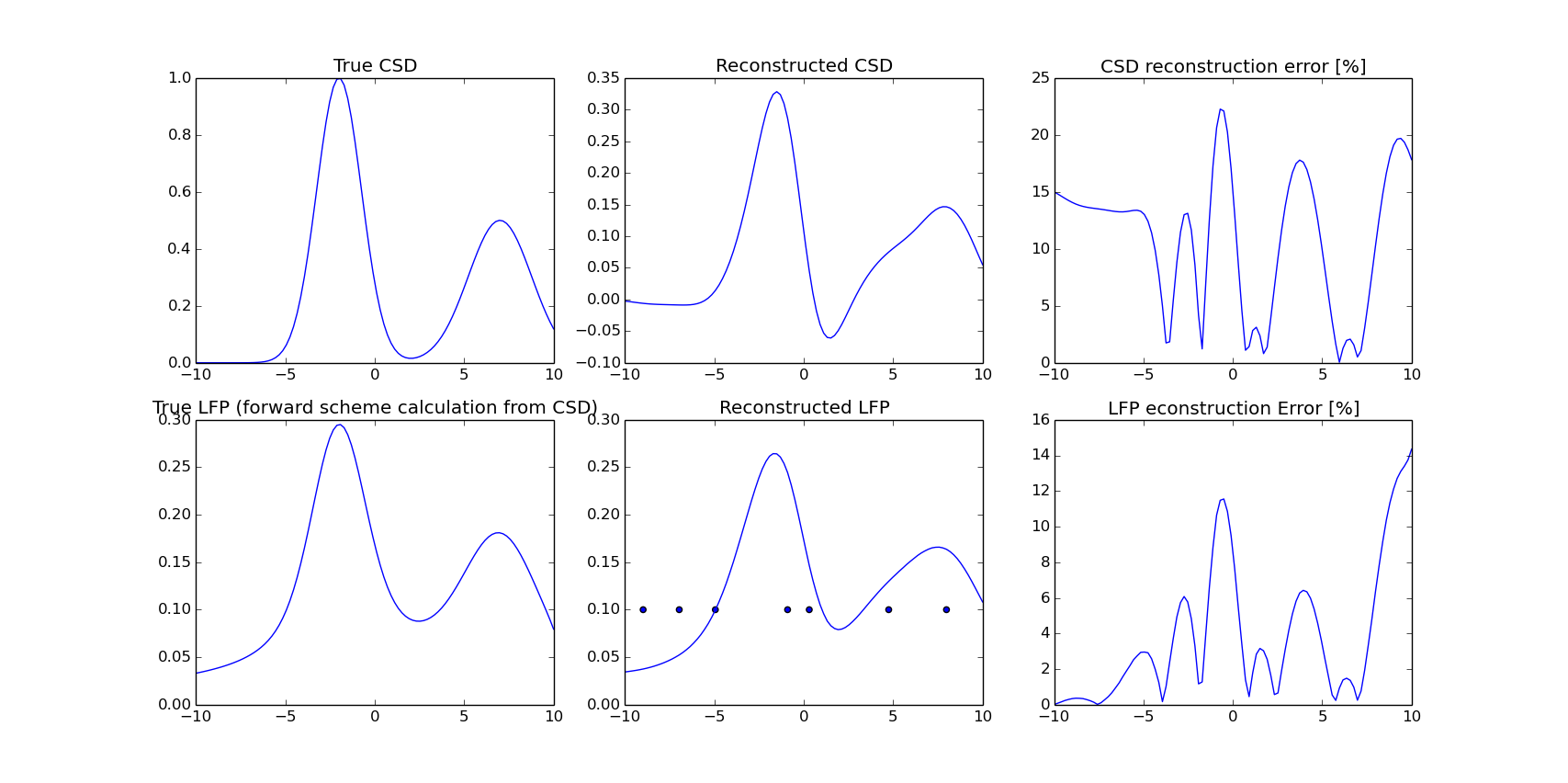
1D reconstruction
x = np.linspace(-10, 10, 100)
true_csd = 1.0 * np.exp(-(x + 2.)**2/(2 * np.pi * 0.5))
true_csd += 0.5 * np.exp(-(x - 7)**2/(2 * np.pi * 1.0))
elec_pos = np.array([-9.0, -6.9, -3.1, -0.3, 2.5, 5.7, 9.3])
params = {'xmin': -10, 'xmax': 10, 'gdX': 0.20}
indx = [5, 15, 25, 45, 51, 73, 89]
compare_with_model_1D(x, true_csd, indx, params)
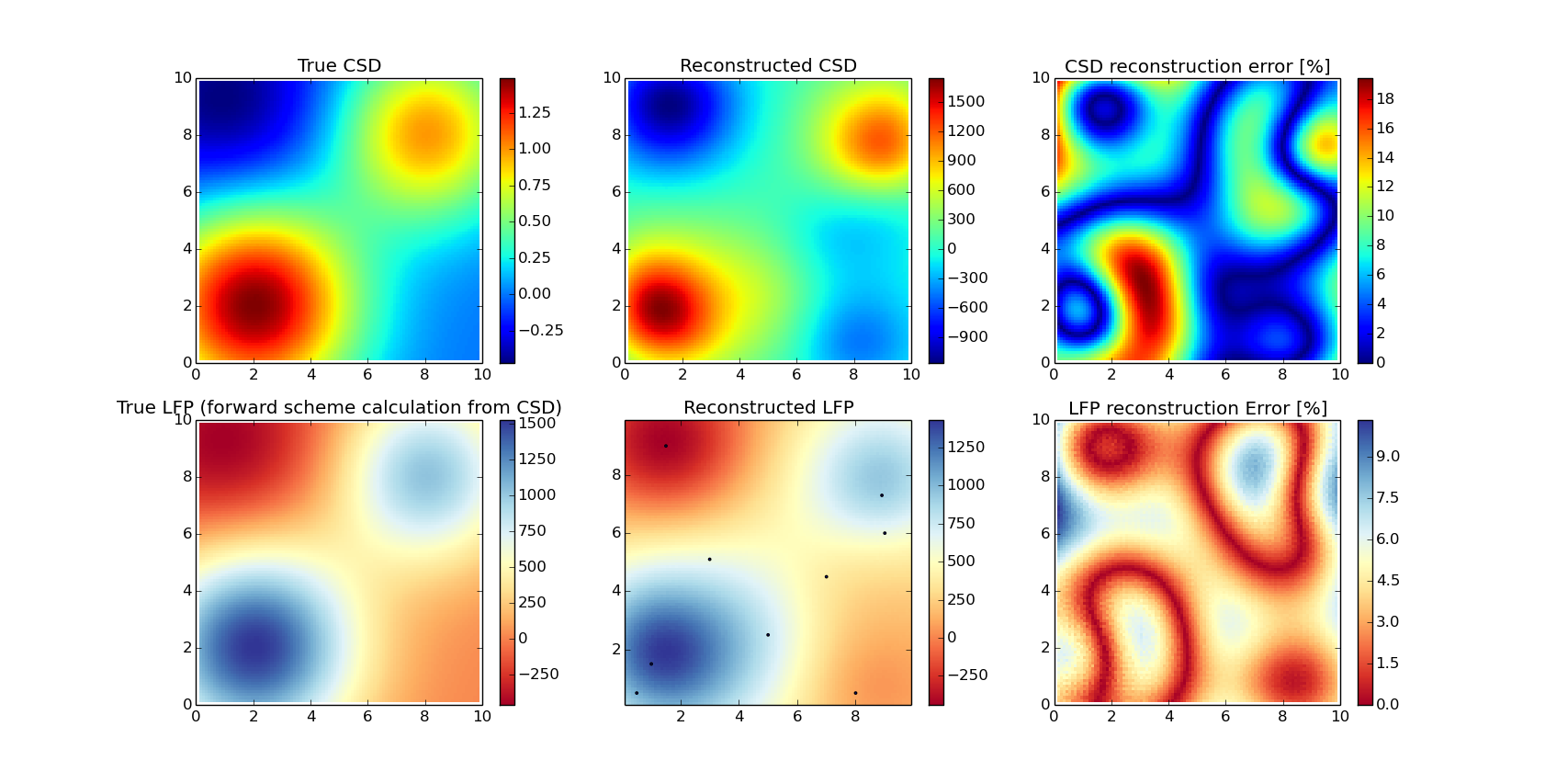
2D reconstruction
xlin = np.linspace(0, 10, 101)
ylin = np.linspace(0, 10, 101)
X, Y = np.meshgrid(xlin, ylin)
true_csd = 1.0 * np.exp(-((X - 8.)**2 + (Y - 8)**2)/(2 * np.pi * 1.5))
true_csd -= 0.5 * np.exp(-((X - 1)**2 + (Y - 9)**2)/(2 * np.pi * 2.0))
true_csd += 1.5 * np.exp(-((X - 2)**2 + (Y - 2)**2)/(2 * np.pi * 2.0))
kcsd_params = {'xmin': 0, 'xmax': 10,
'ymin': 0, 'ymax': 10,
'gdX': 0.10, 'gdY': 0.10,
'h': 0.5}
indx = [[5, 5], [15, 10], [25, 50], [45, 70], [51, 30],
[73, 89], [5, 80], [90, 15], [60,90]]
compare_with_model_2D(X, Y, true_csd, indx, kcsd_params)
3D reconstruction is missing as I haven’t yet found a clear and simple way to display multiple 3D plots.
Time series management
The pykCSD toolbox can now estimate CSD for time recordings and calculate multiple frames. However with many time frames and especially in the 3D case, the memory usage can go very high. This is yet to be optimized.
What’s left:
- Comparing images matlab/python visually (with existing datasets)
- tests on bigger datasets: 100, 1000, 10k electrodes and time frames
- describing workflows for documentation
- package deployment

